Analysis of Rat Kidney Cross-Sections by MS Imaging
MS imaging uses a mass spectrometer to visualize localized biomolecules and metabolites on pathology tissue sections. It is attracting attention as a technology that provides important information for disease detection and treatment.
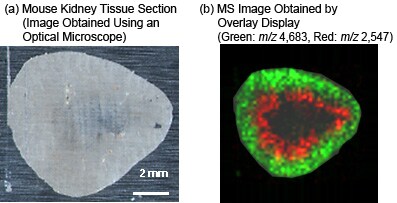
The following shows the results of MS imaging of peptides and proteins identified on a rat kidney tissue section (frozen section) spotted with matrix (sinapic acid) at 200 µm intervals.
The 2D distribution of m/z 2,547 ions and m/z 4,683 ions was color coded on the tissue section and then overlaid to produce a 2D distribution map (b).
MALDI-TOF MS imaging enables visualization of the differentially-expressed molecules of specific mass, such as peptides and proteins, on tissue sections.
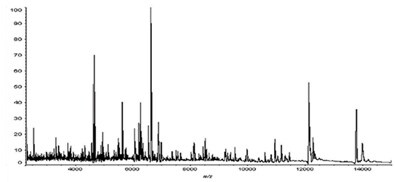
MS Spectrum of Minute Region on Kidney Tissue Section
What Is MALDI-TOF MS Imaging?
MALDI-TOF MS imaging, which uses MALDI-TOF MS, is an emerging technology that allows direct measurements of biomolecules and metabolites on a tissue section, without sample extraction or labeling. The positional information and detected signal intensities can be used to create 2D distributions of target biomolecules. Pretreatment using a CHIP-1000 Chemical Printer to dispense trace, picoliter, levels of enzymes or MALDI-TOF MS matrix allows MALDI-TOF MS imaging with exceptional reproducibility and accuracy.
Features of MALDI-TOF MS Imaging

- The CHIP-1000 Chemical Printer can dispense trace, picoliter, levels of a diverse range of reagents including matrix solutions and enzymes to facilitate MALDI-TOF MS imaging with excellent reproducibility.
- The CHIP-1000 enables MALDI-TOF MS imaging, identification, and structural analysis of diverse molecules, from lipids and low-molecular compounds to peptides and proteins.
- The acquired MS data can be read and analyzed using existing MS imaging software, such as BioMap (http://www.maldi-msi.org/).
- For details on MALDI-TOF MS, click here.


